Visualizing biomolecular electrostatics in virtual reality with UnityMol-APBS
PubDate: Nov 2019
Teams: Pacific Northwest National Laboratory;Université de Paris;PSL Research University;Brown University
Writers: Joseph Laureanti, Juan Brandi, Elvis Offor, David Engel, Robert Rallo, Bojana Ginovska, Xavier Martinez, Marc Baaden, Nathan A. Baker
PDF: Visualizing biomolecular electrostatics in virtual reality with UnityMol-APBS
Abstract
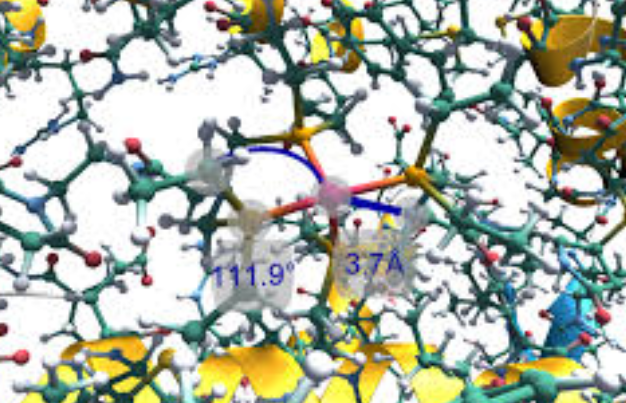
Virtual reality is a powerful tool with the ability to immerse a user within a completely external environment. This immersion is particularly useful when visualizing and analyzing interactions between small organic molecules, molecular inorganic complexes, and biomolecular systems such as redox proteins and enzymes. A common tool used in the biomedical community to analyze such interactions is the Adaptive Poisson-Boltzmann Solver (APBS) software, which was developed to solve the equations of continuum electrostatics for large biomolecular assemblages. Numerous applications exist for using APBS in the biomedical community including analysis of protein ligand interactions and APBS has enjoyed widespread adoption throughout the biomedical community. Currently, typical use of the full APBS toolset is completed via the command line followed by visualization using a variety of two-dimensional external molecular visualization software. This process has inherent limitations: visualization of three-dimensional objects using a two-dimensional interface masks important information within the depth component. Herein, we have developed a single application, UnityMol-APBS, that provides a dual experience where users can utilize the full range of the APBS toolset, without the use of a command line interface, by use of a simple graphical user interface (GUI) for either a standard desktop or immersive virtual reality experience.